Instead of a colonoscopy, a new screen tests poop for colorectal cancer-related bacteria
Testing for the bacterium Parvimonas micra in stool may present an alternative to a colonoscopy
Photo by Janelle Hiroshige on Unsplash
Let's imagine a patient named John. For the past few months, John has had uncomfortable bowel movements, occasional bloody stool, constipation, and lately, unexplained fatigue. His human fecal immunochemical test (FIT), a preliminary test for colorectal cancer, has signaled the presence of a mass of cells in his large intestine. He's been on a liquid diet for the past day. In the evening, he drinks a Gatorade mixed with laxatives. He'll have another in the morning.
He's preparing for a colonoscopy. John is not alone. Colorectal cancer (CRC) is the third most common cancer, with over 1.8 million new cases and 0.9 million deaths worldwide each year. Though the rate of incidence is lower in middle- and low-income countries, the rate of death is higher than in their high-income counterparts. This can be attributed to fewer resources and not widely available adequate health centers in such nations.
The availability of a colonoscopy, the traditional method to screen and diagnose individuals with CRC, in such countries is restricted to tertiary hospitals in cities. Many need to travel long distances to get this inconvenient procedure done. As a result, a sizable portion of at-risk people don't show up for the procedure.
An FIT, which aims to spot hidden red blood cells in feces, is the most accepted non-invasive approach. But it can only detect cancer lesions that are bleeding, which is 35-48% of them. Besides, it is not sensitive enough to detect advanced stages of CRC. Lately, computerized tomography is emerging as an alternative, but it exposes patients to harmful radiation.
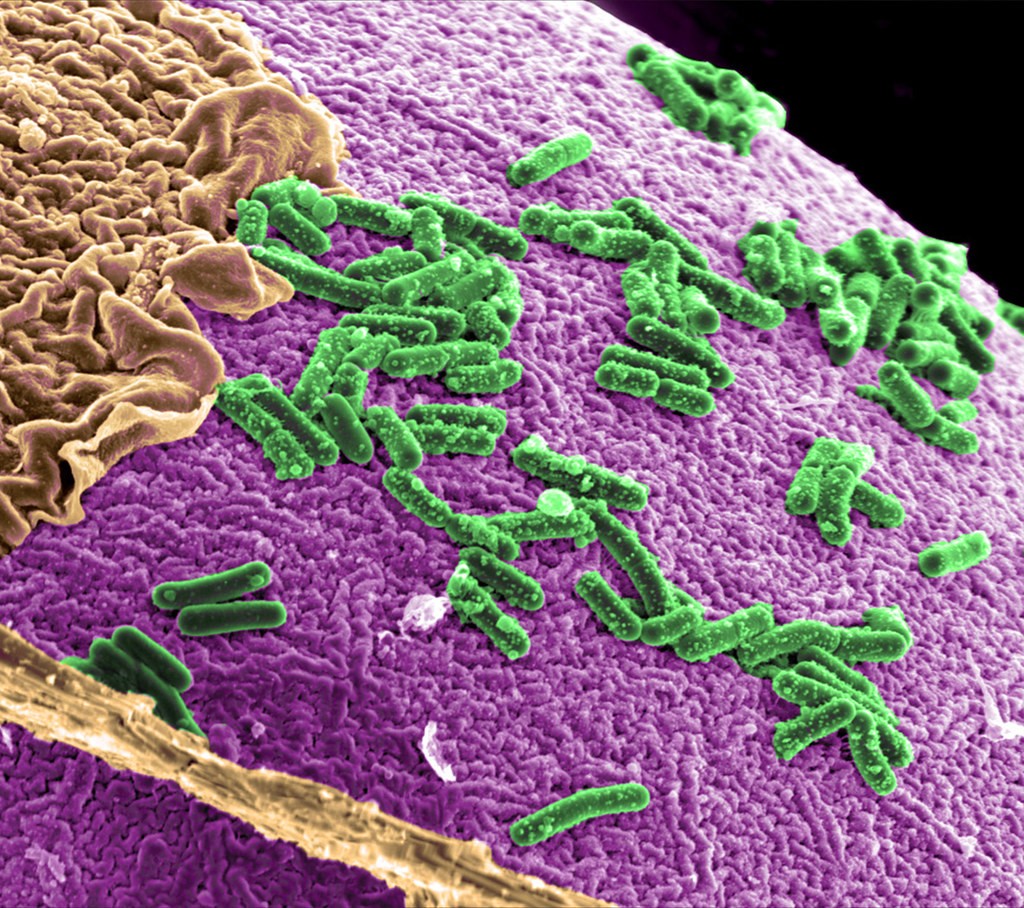
Intestinal microbiota
PNNL via CCSearch. Licensed under CC BY-NC-SA 2.0
In a new study, researchers at Umeå University in Sweden proposed a convenient diagnosis test for colorectal cancer that uses stool samples. They used measurement of quantity of bacterial species called Parvimonas micra in feces as a microbial biomarker of CRC.
In the human body, the large intestine is densely populated with microbes, with around ~1014 bacteria living there. Recent studies have shown that variations in the intestinal microbiota are connected with CRC. In last few years, the microbiome's role in formation and progress of intestinal tumor through DNA damage, inflammation, and immunity modulation has come under scrutiny. For instance, researchers have shown that implanting fecal bacteria obtained from CRC patients causes tumor growth in healthy mice.
P. micra is a usually harmless bacterium that resides in our mouth and intestine, though, under some conditions, it may cause severe gum infection and lung abscesses. Earlier studies have shown that it has a role to play in CRC.
Dietary and lifestyle changes result in creation of an imbalance between the pathogenic bacteria and commensal bacteria. This is sometimes called "dysbiosis" and often results in pathogenic bacterial overgrowth causing various bowel disorders, including CRC.
Three groups of humans were involved in the current experiment. Individuals in one group showed signs of dysbiosis like bowel discomfort and diarrhoea. The second group was of colorectal cancer patients, present in different stages of the disease. The third group was of individuals without dysbiosis complaints and CRC.
The researchers observed an unusually large presence of P. micra in CRC patients compared to non-CRC individuals. Interestingly, neither patients with dysbiosis or non-CRC individuals showed this overgrowth of P. micra. This signifies that in healthy individuals and people with non-CRC dysbiosis, P. micra grows at its usual pace, but that in the presence of a tumor it grows rapidly.
Dysbiosis can be a pre-cancerous stage (but it doesn't have to be). The absence of unusual P. micra growth in dysbiosis shows that the bacteria is not responsible for genesis of tumor lesions. Its high number only in CRC patients indicates that it is an opportunist that begins to grow aggressively, finding a suitable breeding ground offered by the tumor microenvironment.
Hence, the researchers concluded that it is unlikely that P. micra is responsible for the initiation of cancer. Rather, it is a "passenger" who starts multiplying aggressively in the altered niche around the tumor.
The "passenger" name comes from a theory called bacterial passenger-driver model, proposed in 2012. This model describes the role of intestinal microbiota in the onset and progress of CRC. It says that cancer of intestinal cells is initiated by bacteria playing the role of a "driver." Examples of such bacteria are members of the family Enterobacteriaceae and the genus Bacteroides spp. They cause DNA damage and promote abnormal multiplication of epithelial cells in the intestine. The process creates an altered niche that paves the way for outgrowth of opportunistic bacteria referred to as "passengers" like Fusobacterium spp., members of the genus Roseburia, and genus Faecalibacterium.
Furthermore, the present test established a specificity of 87% and a sensitivity of 60%. For a medical test, sensitivity and specificity are two important qualities. Sensitivity and specificity are the ability of a test to correctly classify a person as having a disease or not having a disease, respectively.
The researchers found P. micra often growing together with another passenger bacterial species, Fusobacterium nucleatum, in the CRC microbiota. They also found that a combined test that detected both P. micra and F. nucleatum, along with an FIT has superior performance than each one alone.
This study also found that the number of P. micra is uniform among all stages of cancer — the bacteria blooms in the presence of the tumor and stays high throughout the course of disease. This is significant because it means that this feces-based test can detect late-stage cancer lesions too, where FIT can sometimes fail. Besides, complementing FIT with microbial biomarkers may help to detect non-bleeding tumors, as it doesn’t rely on finding red blood cells in stool.
To cap it all, an efficient stool-based test, such as this, could reduce the diagnostic burden of CRC on patients. A convenient collection of fecal samples, which can be later delivered to designated laboratories, will certainly increase participation from patients, especially in countries with limited resources.
P. micra, in combination with other bacteria biomarkers and FIT, is a suitable candidate for designing a non-invasive screening test for CRC. Moreover, there is a need to create a screening panel consisting of multiple such microbial markers from gut microbiota. This could enhance the overall performance of the panel and may minimize one’s need to go through colonoscopy.
Peer Commentary
Feedback and follow-up from other members of our community
Simon Spichak
Neuroscience
Great piece! Often studies might differ in the way they process poop, leading to differences in their results. Has Parvimonas micra been detected by other studies looking at colorectal cancer? What kind of sequencing did the authors use?
The sensitivity and specificity still remain a little too low for clinical effectiveness. Nonetheless I think this opens up the door to finding more biomarkers for this disease, or even as warning signs to determine which patients might need a colonoscopy and which won’t.
Madeline Barron
Microbiology
University of Michigan
The bacterial passenger-driver model you discuss is new to me and a really interesting framework through which to think about intestinal host-microbe interactions. Even in homeostatic conditions, I imagine we have some key “drivers” that shape the environment to promote the establishment/persistence of passenger species. It’s interesting to think how who is in the driver vs. passenger seat likely depends on host and microbiota-associated factors and varies depending on gut environmental context (e.g. health vs disease).
While I know this wasn’t the major takeaway from the study (which was super cool), I’m always interested in the bug side of things— did the researchers speculate about how the tumor environment specifically shapes a niche for P.micra and other bacterial passengers? Do they think these bacteria might worsen disease? I wonder what these findings could mean not only from a screening perspective, but also a treatment one (given interest in therapies that re-shape the gut microbiota for a number of diseases).
Namrata Iyer
Microbiology
Brown University
There’s a lot of work being done to prove a causative relationship between microbes and disease. But it’s useful to think that even those microbes that don’t cause disease, are happy to take capitalize on the altered environment. Diagnostics is an exciting application for microbiome field; especially for a disease like CRC where the current diagnostic gold standard is expensive, invasive and highly uncomfortable. I wonder what geographic cohort the study was performed on. Diet and lifestyle cause healthy microbiome to vary significantly across countries. It’d be interesting to see if the same bacteria tend to overgrow in CRC patients from different countries, which will affect the kind of diagnostics that need to be done. Sequencing stool samples, though non -invasive, is still not cheap or easily accessible in developing countries. Hope the researchers can build on this to make it easier to transition to a clinical scenario. Thanks for highlighting this work!
Luke Whitcomb
Physiology, Biomedical Sciences
Colorado State University
I especially appreciated your discussion of how colonoscopies, though the diagnostic gold-standard for CRC, may be inaccessible or dissuasive for some patients populations. I think a lot about health care equity here. The average cost of a colonoscopy in the USA is around $3,081 for those not covered by health insurance (and 14.7% of US adults aged 18-64 were estimated to be uninsured in 2019). Even with insurance, patient can expect to pay between zero to $1000, depending on their type of insurance coverage. In contrast, FIT tests can cost exponentially less. Combined with P. micra detection, such methods could be invaluable means of increasing patient access to colon cancer screenings. Thanks for sharing these new diagnostic advancements!