Scientists are growing mini-intestines that stretch and bulge just like the real thing
This could lead to developments in personalized medicine and treatment of chronic intestinal diseases
Portrait of Samuel D. Gross - Thomas Eakins
Researchers worldwide are attempting to grow a variety of human organ models in the lab. It sounds unreal, but scientists in Boston are even growing miniature human intestine models in the laboratory.
Modeling organs could help scientists study everything from how different cells interact to the mechanisms of how diseases develop, providing new insights into treatments for a variety of illnesses and injuries. These kinds of ambitious projects are motivated in part by the problem of reproducing data, a crisis that has been plaguing researchers, doctors, and drug companies for the past decade.
A general tenet of science is that if an experiment is repeated precisely, its conclusions should be the same. But in fact, a survey by Nature asked 1,500 researchers whether or not they believed that science was experiencing a crisis of reproducing results, and 52 percent responded, "Yes, a significant crisis." Specifically, over 50 percent of researchers responded that they have been unable to reproduce their own research, and 70 percent have been unable to reproduce a colleagues'. This crisis has high societal costs. It has been estimated that in the US, 28 billion dollars are spent per year on pre-clinical research that cannot be reproduced.
Data consistency is especially important — and difficult to obtain — when it comes to complex systems like the intestine. This is due, in part, to the community of microbes that exist inside the intestine, which can vary significantly from person to person. Known as the microbiota, these bacteria are essential to human health and disease.
Unfortunately, it has proved challenging to study the microbiome in humans. Scientists must rely on people collecting samples of their feces, and keeping these samples in their fridge until they deliver the fecal sample to a laboratory, ideally within 24 to 48 hours of collection. Many studies use rodent models as a surrogate instead, but unfortunately, rodents lack some types of bacteria that are linked to healthy human microbiomes. They're far from perfect substitutes. And microbiota quickly change based on diet, water, and location — even moving mice between facilities can cause short term changes in their microbiota.
For example, a group of scientists at the University of California at Davis decided to investigate whether microbiota impacted how genetically-related mice got sick. Andreas Bäulmer and his colleagues found that the different microbiomes led to different likelihoods of viral infection. Although this study looked at viral susceptibility, their finding that mice purchased from different companies have different microbiota could potentially influence the results of other studies. But despite these limitations, animal models have thus far been the best system we have for most disease research.
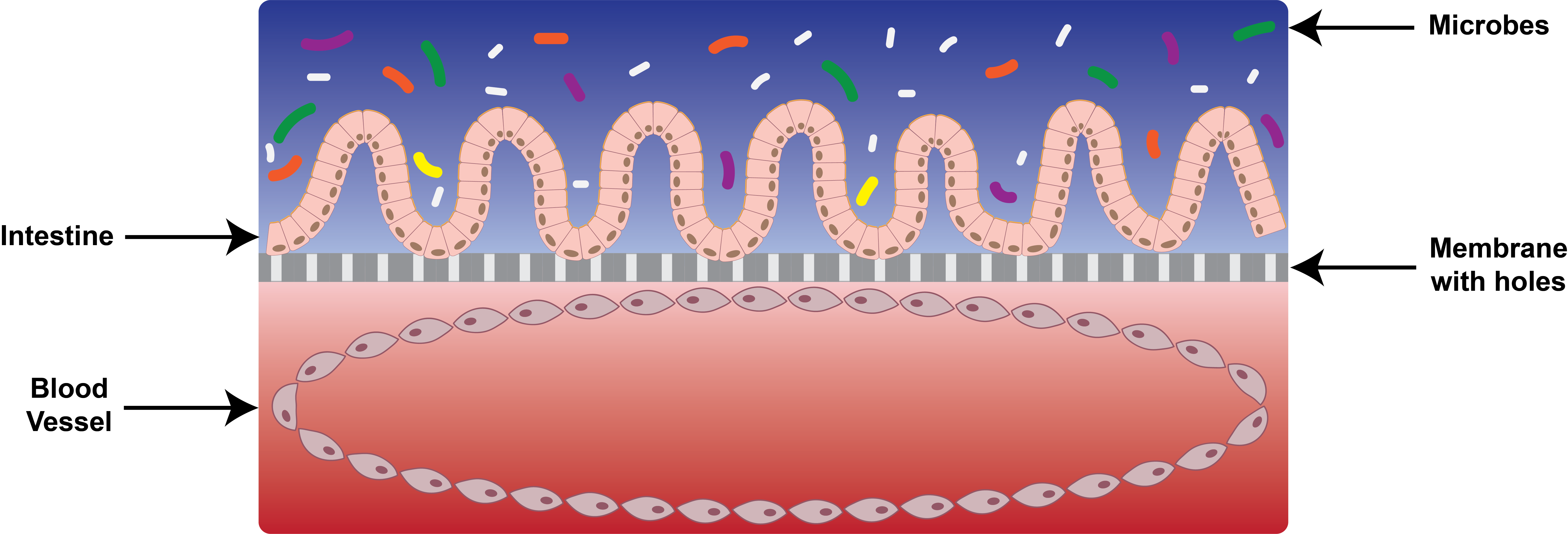
Cartoonized cross-section of the intestine-on-a-chip
Illustration by Juliann Tefft
As it has become increasingly clear that mouse models are insufficient, Donald Ingber, a professor of bioengineering at Harvard, is finding a way to develop human organ models in his lab as alternative ways to study human disease. This was motivated by the discrepant environments cells experienced when grown the laboratory compared to how they exist naturally in the body. In a laboratory, cells are grown on very stiff plastics, and are not experiencing dynamic forces. This is quite different from the body, where organs are constantly moved and stretched. Could growing cells on flexible plastics bridge the gap between the lab and real life, and improve our ability to model organs in the lab?
So in 2010, Ingber took a thin flexible plastic and coated it with lung cells. Stretching the lung cells changed how small particles moved through the lung, starting a flow of publications which incorporate forces with other organ models. The next target — the intestine. Intestinal cells have to survive several different forces: food stretches the walls of the intestine in multiple directions and rubs along the intestinal walls as it flows through the digestive tract. Also, the intestine experiences two distinct oxygen environments, one on the outside of the intestine, where a system of blood vessels brings fresh oxygen to the tissues, and another on the inside where oxygen levels are incredibly low. So a model of the intestine needs to be able to be repeatedly stretched, handle fluids, and survive both high and low oxygen levels.
To do this, Ingber started with a 3D piece of flexible plastic about the size of a fingernail. Inside, two channels are separated by a thin membrane, with holes large enough for fluid and molecules to flow through, but too small for cells. In the low-oxygen channel, intestinal cells are attached to the permeable membrane. On the opposite side of the membrane, blood vessel cells attach to the walls, forming a cylindrical blood vessel with higher oxygen levels. Meanwhile, intestinal motion is simulated by repeatedly stretching and compressing the plastic chip.
Initial versions found that this stretching helped mimic a real human intestine. Ingber's lab also incorporated individual bacteria onto the chip, while later work went further to add immune cells and commercial probiotics. In their most recent version, published in Nature Biomedical Engineering in May 2019, Ingber and his colleagues demonstrated that their improved chip can sustain gut bacteria which grow only in negligible levels of oxygen.

Studying the human gut microbiome just got easier, thanks to the intestine-on-a-chip technology
This study explored the potential of the chip as a model for developing personalized treatments. Ingber and his collaborators tested the chip with cells from patients at Boston Children's Hospital. The researchers isolated intestinal cells from a patient who had undergone surgery, and microbiota from baby feces, and added them to the intestine chip. These cells formed a healthy multi-layered intestine, and survived for five days. Ultimately, their goal is to connect many different mimicked organ 'chips' in order to predict how drugs will affect human bodies.
Although we're still a ways off from being able to use this technology at a doctors' office, these are exciting steps toward personalized medicine. In the future, intestinal models like these could help us better understand complex diseases like Crohn's disease, an intestinal disease that can be triggered at any age. Cells from people with Crohn’s could be used to investigate why some people develop the disease and others do not. Although Crohn's disease effects 700,000 Americans, animal models have only been able to mimic certain aspects of the disease.
Organizations like the Mayo Clinic are developing cell biobanks, where samples from healthy people and those with Crohn's disease are frozen for future research. Companies with these biorepositories could be pioneers in using humanized organ-on-a-chip models for personalized medicine. In the future, pharmaceutical companies could use these models to test drugs before beginning expensive clinical trials, potentially decreasing the cost and increasing successes of drugs in clinical trials. The cost of manufacturing the plastic chip is low (although the process of adding cells is nuanced and expensive). But as the model's promise grows, several companies have already been founded to help grow the technology, and hope to improve the reproducibility of organ research between labs by making these chips commercially available.
Peer Commentary
Feedback and follow-up from other members of our community
Marnie Willman
Virology
University of Manitoba Bannatyne, National Microbiology Laboratory
I still find this concept fascinating. It’s great to do animal and cell line research, but when it comes to frontline medicine like determining which drugs will be best for a patient, nothing works as well as the patients own cells! Everything else we’ve had so far to mimic human physiology is a far cry from the real thing, but it’s the best we had at the time.
I hope to see more of this research take the place of microbiome bacterial work and animal research, as it is a more accurate representation of human anatomy and physiology. What used to read as science fiction, as eloquently put in the article, is finally becoming a reality. What a breath of relief for people needing personalized treatments for various ailments.
My only moment of doubt is considering how the immediate environment compared to the entire organism functions. We now know that the intestinal microbiome affects all areas of physiology, and likely vice versa. So, how will scientists ensure that the microenvironment seen in artificial mini-intestine organoids accurately mimics the entire human body’s interactions? Time will tell! Great piece.
Alaa Ahmed
University of Georgia
This is a great read! I find this whole progress in how we’re looking at biological processes impressive, through individual organ models. Like discussed in the piece, there are a lot of variables involved in organ behavior in its natural biological environment. As a microbiologist, I liked the introduction of individual bacteria to the chips, but I’m also kind of curious as to how they’ll address the presence of diverse communities, rather than single species (if they haven’t already).
The kicker was also great, providing an outlook on the future of the technology.
You hit on a good point - sustaining complex communities is certainly important. I didn’t go into it in much detail in this article, but in the 2019 Nature BME paper they do use complex microbiota samples. In fact, two of the figures investigate how a few different bacteria within this microbiota change abundance over the course of 3 days. My interpretation is that there’s a relatively significant shift in the microbiota’s composition during the first day of culturing in the chip, but the profiles are relatively stable between days 1 and 3 of culture. I think this is useful to see and leaves room for improvement in future work so that the microbiome is truly representative of human samples.
Another caveat is that the focus has been on demonstrating diverse bacteria rather than non-bacterial components of the microbiome. I think this is generally consistent the standard of the field at this point, but ultimately a full microbiota should also include microbial eukaryotes!
Max G. Levy
Science and Health Journalism, Chemical Engineering
This is so cool! And such a great example of the systems we can look at on a chip. Based on what I’ve read, this tech can be really useful for zooming in on super specific biological happenings. I wonder how long it will take before we see FDA approval of a drug that has been tested on a chip since Ingber’s start-up partners with the FDA!
Thanks! I agree, it will be really interesting to see the outcome of the FDA testing these chips. As you probably know, currently new drugs must be tested pre-clinically in animals before human clinical trials are possible, so these tests are really important. I imagine it will be a while before animal trials are phased out, but this is a really important step to evaluate the consistency of these models!
Tara Fernandez
Cell Biology
Really well written Juliann! As engineered tissues become increasingly complex, their potential for becoming sophisticated research tools and diagnostic platforms really skyrockets. One concept I’ve been exploring recently is scientists’ ability to recreate not just single tissue types in isolation, but miniature networks of organ systems.
This year, researchers published a world-first study in Nature, premiering a new method for turning stem cells into 3-dimensional models of not one, but three interconnected organs: the liver, bile duct, and pancreas. Like Ingber’s intestinal culture system, these organoids are paving the way for the next-generation of in vitro models and, in the future, perhaps even on-demand lab-grown organs for transplantation. Exciting stuff!