Scientists are testing for coronavirus with genetic "fishing hooks"
Molecular "baits" also make testing for the virus safer for lab personnel
John Crawford, National Cancer Institute, NIH
"These baits act as fishing hooks; we can throw these hooks in and physically pull out the virus of interest."
The virus that Jalees Nasir is interested in is SARS-CoV-2, the novel coronavirus responsible for the COVID-19 outbreak that started in China, in late 2019. At the time that this article was written, there are over 716,000 confirmed cases and 33,000 deaths associated with the virus.
When a person walks into a hospital with symptoms related to COVID-19, one of the first things a doctor might do, in an ideal world, is collect a sputum sample by asking them patient to cough into a cup.
"Say, you have a patient sample," says Nasir, "you're likely to have a combination of human material and bacterial material – you can have viral material as well, but the proportion of the virus will be far smaller than that of human or bacterial."
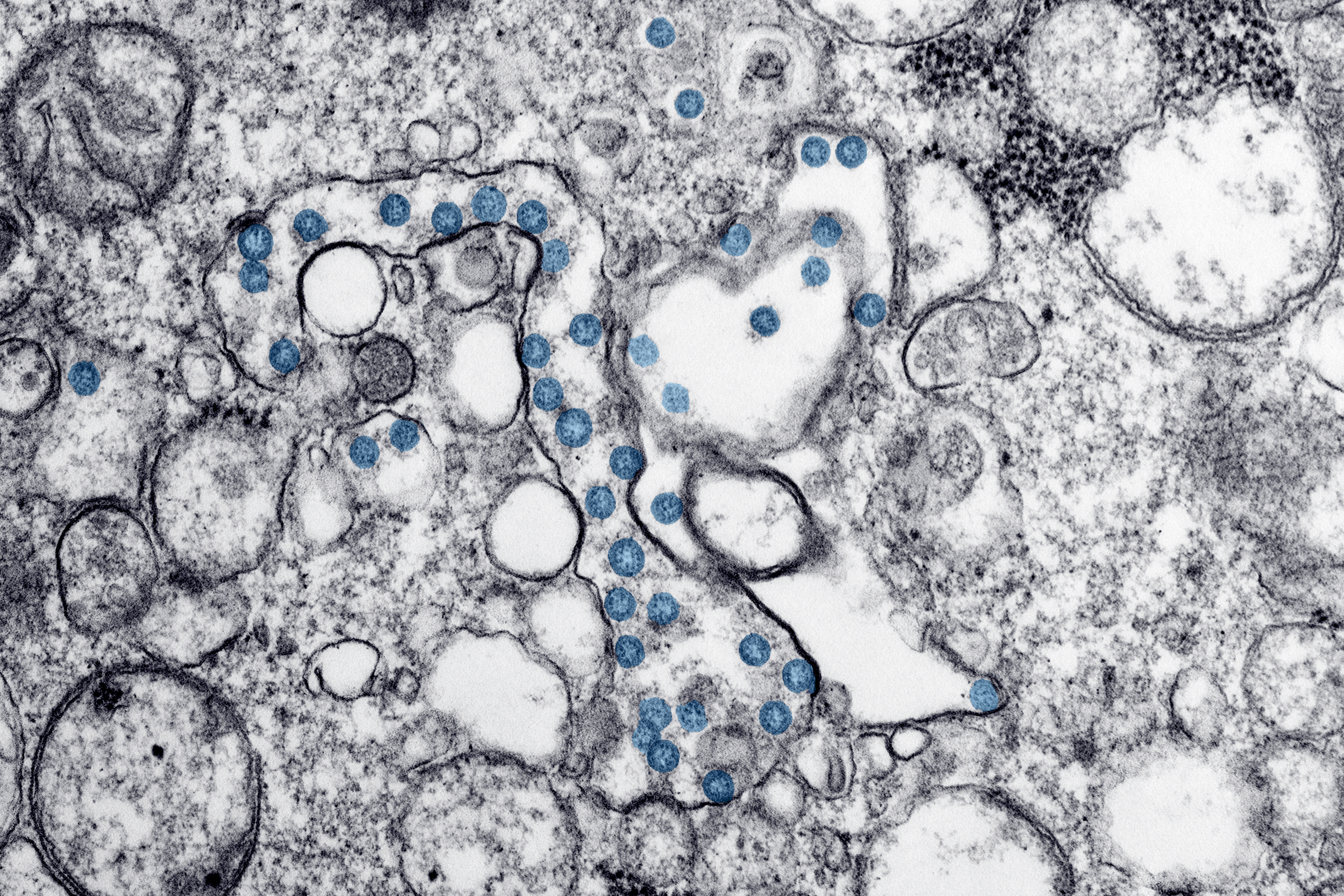
A microscopic image of coronavirus particles, colored blue, in a patient sample. The virus's genetic material can be seen as little black dots within the blue particles.
Hannah A Bullock, Azaibi Tamin, via CDC
In a nutshell, it's a biological soup. To study the viral genome, a microbiologist has to grow more virus to get a large enough sample. To do that, the virus has to be separated from everything else. This kind of work is designated Biosafety Containment Level III — the second most dangerous category for hazardous biological laboratory work. This is where scientists work with microbes that can be easily spread through respiratory transmission. Isolating and growing the virus is not only expensive, but it also increases the risk of exposure for anyone working with the viruses, in comparison to working with the relatively small amounts of virus in patient samples. Another concern is that the virus could further mutate during its time growing in the lab, and the final genomic sequence could be different from what was in the patient.
An alternative is to take that complex sputum sample, and sequence the genetic code of everything in that complex sputum sample. But you might not even get any viral RNA or DNA because of how much other stuff is in there. This is known as metagenomic sequencing, and is typically even more expensive to run.
To fix this problem, scientists at McMaster University in Canada have developed a set of molecular probes, or "fish hooks" capable of pulling the coronavirus from those complex patient samples. This method, commonly used for sequences of ancient samples, involves "fishing out" the desired target molecules with probes that specifically attract viral genomes.
Being able to pull out and sequence the virus in a safe and cost-efficient manner is important to study viral transmission and evolution.
"We actually want to examine the differences that could potentially occur in transmission between Patient A and Patient B," says Nasir. "We need to start looking beyond that diagnosis aspect. We don’t want to just say, 'We have SARS-CoV-2,' but that 'We have this specific strain of SARS-CoV-2 from Patient B,' where it might have mutated during the transmission. We need to see the specific differences."
There are three layers to the epidemiological response surrounding an outbreak. The first involves the front-line doctors and clinicians who diagnose and treat the people. The second layer, which is also the primary focus of the research community, involves finding an immediate solution — vaccines, cures, and treatments. The third layer is about surveilling the outbreak.
How fast is the virus spreading? Is it mutating and evolving as it moves from one patient to another? Are the viral strains in Italy the same as the ones in China?
These are exactly the kinds of questions that scientists working at this level are trying to answer. And, needless to say, these answers matter when formulating a global response to an outbreak. However, these questions have been difficult, risky, and expensive to answer in the past, because it requires an amount of lab work and genomic sequencing that isn't feasible for every single case.
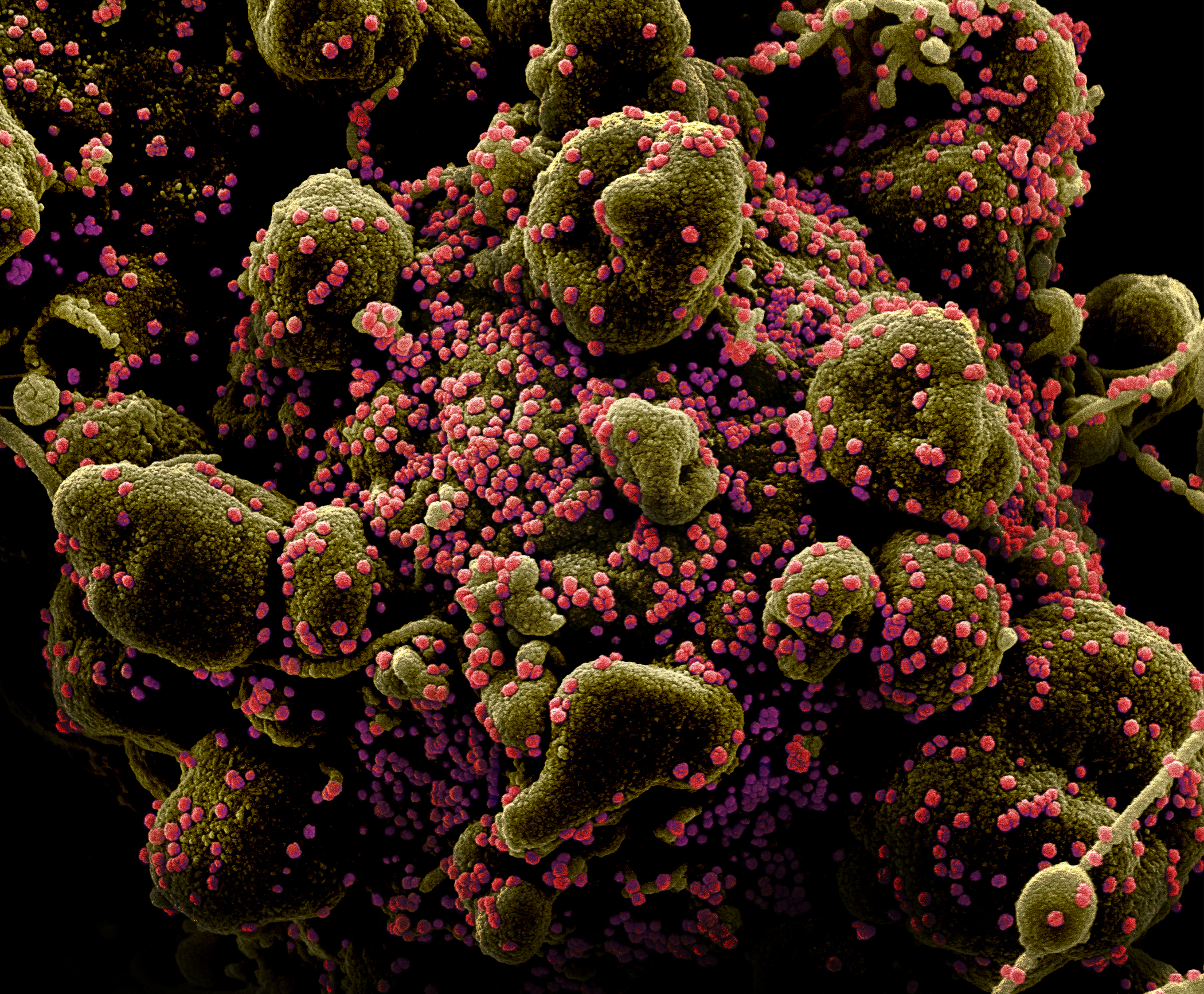
A picture of SARS-CoV-2, the novel coronavirus, particles on the surface of a human cell undergoing self-destruction.
"In this case, we have hopefully made these baits as specific as possible to SARS-CoV-2, so that the virus of interest we end up pulling is, in fact, the novel coronavirus," says Nasir.
As a PhD student in Andrew McArthur's lab, Nasir had been working on creating molecular probes for respiratory viruses for over a year.
"When the outbreak started, I was more or less still in the design phase. At that time, I thought I should double down." Nasir immediately started comparing the novel coronavirus to the pre-existing probes he had made for other respiratory viruses. "I walked into Andrew’s office and I said, 'We could potentially hit this'."
As soon as they validated the "bait" design, McArthur and Nasir wrote up their findings and shared it with the research community.
"We could go the traditional academic route, and wait for the samples and find a collaborator, and then it wouldn’t be useful. This is crowdsourcing the science," says McArthur. "We did the first part, which is the design, and it’s what we’re good at. And now we crowdsource the validation. Like any assay, it might have to be tweaked once people start testing it."
A similar bait set was also released by an independent research group in Arbor Biosciences, further validating these results. The company is also providing their 2019-nCoV hybridization capture kits to researchers around the world without any cost.
Since molecular probes would extract only the viral material from the entire patient sample, this would reduce the additional costs from the alternative methods — such as sequencing the entire patient sample, or growing the viral sample in lab. This process could be made even faster by using sequencing technologies like the Oxford Nanopore, which is a portable sequencer that can sequence longer genomic strands at a time. Used together with more traditional Illumina sequencing, this could give researcher high quality data to compare even single base-pair mutations in the virus. High quality genomic data is essential when it comes to developing a vaccine.
"I think any discussion of vaccine has to tempered with the fact that we have not yet been successful with SARS, so we could have the same poor success rate with this virus," McArthur says, "But, if this particular virus was much more workable for a vaccine, we would want to know what the antigen was, and which gene encodes the antigen. And from the sequencing, we want to know how it’s evolving. We want to know if it has the capacity to out-evolve the vaccine, or whether you’re building a vaccine for a site that’s not likely to evolve, like the measles vaccine."